
The BioEditor allows you to create Bio Sequence Table (.bst) files from standard DNA and Protein data sequences that are freely available on the Internet. Two excellent free sources of genetic sequences are the Swiss Protein Data Bank and the NIH GenBank.
The job of the BioEditor is actually quite simple. It separates the biological sequence data from the source text file, and it converts the data into a numerical table format that can be used by Bio Sequencer modules. Although there are a number of controls in the BioEditor, most of them are for delimiting secondary structure features such as turns, alpha helixes, and beta sheets - which can be used to trigger musical changes or events. For a simple, direct translation of the protein or DNA data, you need only to read in the file and set markers for the beginning and end of the data.
This page documents the controls and functions of the BioEditor; please see the BioEditor Tutorial for an example of how to use it to prepare sequences from the thousands of data files freely available on the Internet.

There are three parts to the BioEditor: the text window, the tool bar across the top, and the list of Bio Sequencer table slots on the right. The text window initially displays the raw text of the source data file, and allows you to edit it as you would with any text editor. It also can display the translated text in either of single letter codes for amino acids, or the corresponding DNA codons (see table).
The Bio Sequencer Table is a binary data file that is formatted so the Bio Sequencer modules can access the DNA, protein, and structure data in real time to produce music from the original text based source file.
When you run the BioEditor, the first thing it does is to load the last table file accessed. The first time it is run, this will be the default BioEditor table file from the Data folder.
The table file is a list of translated codes and their source file names, but not the source files themselves. There are 24 slots in the table, numbered 0-23, which corresponds to the "Seq#" parameter inputs in the BioSequencer modules. File names of any of the 24 slots that have data in them can be seen in the corresponding slot to the right of the editor window.
When BioEditor loads a table file, it searches through the list of file names held in table file it loads, looking for the original source files. If a source file is not found in the location stored in the table, BioEditor looks in its default source directory (which can be set from the Options menu) for the file. If the source file is found in either its original location or in the default source file directory, the label is colored blue, if only the translated table data is found the label remains gray. A a gray label means a source text file will be reconstructed from the stored binary data, but the true, original source is not available.
When you have made changes to the source file, the label text will turn red. The characters P, D, and X just after the slot numbers indicate whether the translation is from Protein source, DNA source, or has not been translated yet.
Click on any of the names in the 0-23 table slots to the right of the edit window, and the corresponding source file will be loaded into the edit window. If there is no source file found (the label is gray, not blue), the sequencer data is back translated. If you save this back translation, the editor will treat it as a source file from that point on. If you click on a blank slot, you will get a blank page that you can manually enter data into. All the standard Notepad editing controls - copy, paste, select all, etc. - are available by right clicking within the edit window.
You can load a new source file with the toolbar Open Source icon, the first one on the left, or the Files menu equivalent. The editor will search for the first empty slot in the table, and load it into that slot. If there are no empty slots, the file will not open and you will get a message asking you to free up a slot.
File extensions of .txt, .pro, and .dna are recognized. The .pro and .dna extensions are intended to make it easier to recognize whether the source is a DNA codon file or an amino acid (protein) single letter code file. However the BioEditor does not distinguish among file extensions - it will accept any filename and extension you give it; and it determines whether you are translating DNA codons or protein amino acids by the context of the data itself.
Once the file is loaded into the editor window, the only edit you must do is delineate the actual code part of the text with tilde (~) marks. The editor will ignore all text outside of the tilde marks, and it will assume any text within the marks is valid data.
You can insert the tilde data markers simply by moving the cursor to the beginning of the data, and inserting the character with the keyboard; or you may highlight the data text, and select the "Start/End Marker" command from either the toolbar button with the tilde label, or the "Flags" menu equivalent.
To translate the delineated code text into Bio Sequencer format, click on either the Translate to Protein or Translate to DNA toolbar buttons, or Edit menu equivalents. These will display the translated text, and they also insert the translated numerical sequence data into the Bio Sequencer table, where the module can access it.
Toolbar: ![]()
Menu: Open Source, Save Source, Save Source As, Revert
Menu Only (no tool bar buttons): New Table, Open Table, Save Table As
Open Source opens a Protein or DNA text source file for editing and translating.
Save Source and Save Source As saves the source text currently displayed in the editor window. It is important to realize that you are only saving the source text, not the translated bio sequence data (described below). Saving the text does not translate it into a sequence - for that you need to select one of the two translation buttons, described below.
You can select Save Source As any time you are viewing source text in the editor window, however Save Source will be grayed out and unavailable unless editing changes have been made in the text. When editing changes are made the slot name will turn red; and if you exit or select another slot you will first be asked to save the current text. If you do not save, the changes are discarded and the next time you open the slot, the original text is displayed.
Revert reloads the current file: the edit window is reloaded with the original source text. The button acts immediately without asking if you want to save changed text. If you hit the button by mistake, use Undo to restore the edited text.
New Table creates a new, empty table. If the current slot has editing changes you will be asked to save it, and if changes were made to the current table you will be asked to save that.
Open Table pops up a dialog box to open an existing table file (.bst).
Save Table As pops up a save dialog box to save the current table (.bst) file with the option to rename it or to save it in a different directory.
Menu Only: Print
Print invokes the Print dialog to print the currently viewed text. This can be either source text or the translated text.
ToolBar: ![]()
Menu: Copy From Previous Slot, Undo
Menu Only: Erase Slot
Copy from Previous Slot is a convenient method of cloning an original source file to make a different set of editing marks. To clone an existing source file, visit it by clicking on its label, then click on an empty slot and select the Copy from Previous Slot button. The new slot will be named "unnamed;" use Save As to give it a new name.
Undo is a one level undo of the last change made to the Source text. It swaps the current page with the last page changed, so you can undo Undo, etc. Typing changes are not saved by Undo, however you can recover from them with the right-click Undo.
Erase Slot clears the currently selected slot from the Bio Table (but it does not erase the Source file itself). If there have been editing changes to the Source file, you will be asked to save it before the slot is cleared.
ToolBar: ![]()
Menu: Show Flags in Translation, View Source, Translate to DNA, Translate to Protein
Show Flags in Translation switches between a display of the translated data alone, or the data with the flag marks included. Flag marks are set by the Flags menu or buttons, described below. This button has no effect on whether the flags are encoded - they are always encoded if they are in the source text - it only determines whether or not they are seen in the display view.
View Source puts the source text into the editing window; Translate to DNA and Translate to Protein translates the source text into binary data that represents just the selected DNA or protein sequence, and displays the translation in the text window.
When either Translate to Protein or Translate to DNA is selected, most of the other buttons will be disabled. You can use the editor to make changes in the text, but it will not be saved as this is simply the translation of the data that has been encoded from the source text. To effect an actual change, you must go back to the View Source mode, and make the appropriate change in the source text. However if you want to use the result of a translation view in another source text slot, you can use the right click edit pop up menu to select the text for copy and paste.
ToolBar: ![]()
Menu: Locate Features, Find/Trim Flags, Delete Flags
These three buttons make it easier for you to find and mark areas of interest in the source code text. Their use is completely optional, but if you start marking some of the secondary structures that are given in the source text as numeric locations, these buttons can make the task considerably less tedious and more accurate.
Locate Features (binocular icon) is probably the most useful. With it, you only need to highlight a set of two numbers in the features area of a sequence file, then press the button. It will jump down to the code area and highlight the codes with those number addresses.
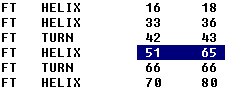
In the example above, the helix at the residue numbers 51-65 is highlighted. No need to be accurate about the highlighting, as long as the numbers are included and there are two of them. At this point, clicking on the Locate Features button will drop you down to the code text, and highlight the inclusive residues:

Note the tilde character (~) has been inserted at the beginning of the code sequence. This must be there for Locate Features to work, as it starts looking for valid codes only after it first finds the tilde start character.
At this point you are likely to click on one of the Flag marker buttons, described below, to mark the area, and then you will need to go back to the (often rather lengthy) feature list - at which point is very easy to forget which feature you just marked. Locate Features makes things a little easier by remembering the last feature list location, and by clicking on it now while you are in the code area it will return the cursor and the highlight to exactly where you started. At this point you may want to enclose the features you just marked with the same character marker you used in the code area, just as a reminder. Just click on the Flag marker button, and the job is done.
Find/Trim Flags finds a matching Flag marker character, and optionally deletes the set. For example if you have text delimited by a set of angle brackets, position the cursor in front of the first (left) bracket, click on the Find/Trim button, and the text from the left bracket to the right bracket will be highlighted. Click on Find/Trim a second time (while the text is highlighted) and the left and right brackets will be deleted.
The rule is: if no text is highlighted when you click on Find/Trim, it looks for the matching marker and highlights the area between the markers; if text is highlighted when you click on it, it trims the markers. The button actually deletes the two end characters from any highlighted text selection, so be sure the markers you want to delete are at the start and end of the highlight.
Delete Flags (trashcan icon) deletes all of a particular Flag marker within the highlighted text. To use it, highlight the text area you want to delete all of a Flag set from. Then click on the button, and it will stay depressed. Now, when you pressed on any of the Flag Marker buttons, all of that Flag set within the highlighted area will be deleted. Clicking on the Start/End marker (tilde), is a special case: it causes all marks within the highlighted selection to be deleted.
Menu: Insert Marker <>, .. (), .. [], .. {}, .. !|, .. @#, .. $%
The group of 7 buttons starting with angle brackets are the Flag Marker buttons. Their purpose is to insert a set of markers into the code text that can be read by bio sequencer programs. It is completely arbitrary how you use these markers, however typically they are used to mark structures in the bio code such as helixes, turns, sheets, transmembrane domains, etc. You can also use them to mark off repeating or similar sections. Whatever you want. There are 7 sets of them. Put the left marker before the position you want to flag, and put the right marker after the position you want to flag. The flags will show up in the MIDI software as On for the duration of that particular marker in the sequence. That is, the flag goes to On when the left marker is detected, and it goes to Off when the end marker is detected.
You are free to manually insert the markers into the text, but the Marker buttons make it a little easier: highlight the code text you want to delineate and click on the marker button you want to use, and a the marks will be inserted at the beginning and end of the highlighted selection.
ToolBar: ![]()
Menu: Start/End Marker, Include Exons, Exclude Introns
These three Markers, starting with the all-important tilde (~) do not show up in the translated sequence as do the Flag markers above. They are instead used to tell the translator which code text to include and which to ignore.
The Start/End Marker (~) tells the translator which text to process. It ignores all text before the first tilde character, and it stops processing when it gets to the 2nd tilde or to the end of the file.
You can insert the tilde character(s) manually, or you can use the button. When you use the button, the entire document is scanned, and any existing tilde characters are removed, then a pair of tildes are inserted at the start and end of the highlighted selection.
The Include Exons and Exclude Introns markers use the same characters - a colon (:) to exclude and a semicolon (;) to include. The translation scanner ignores all text after scanning a colon until it comes to a semicolon - which then starts processing text again. Since the default is to start scanning text after the first tilde character, the semicolon character is not needed unless a colon has been used to turn off scanning.
The two buttons are (; :) to mark text for inclusion (called "exons"), and (: ;) to mark for exclusion (called "introns"). When you mark for inclusion the first colon at the start of the code is not needed, as the scanner assumes that exclusion up to the first inclusion mark (;) is implied.
Menu Only: Change Fonts, Source Look-up Directory, Update Table Source
Change Fonts brings up the Windows Change Font dialog. Only fixed fonts are shown.
Source Look-up Directory brings up a Windows dialog to select a directory, to set the directory that BioEditor will look for any source files that are not found in the location stored in the Table file. You can set any directory you want for this source look up area, but generally it is best to use the same area. The default is the Source directory in the BioEditor program directory.
When BioEditor is unable to find a source file in its original location, but does find it in the default source look-up directory, it will automatically change the file location in the copy of the .bst Table file it keeps in memory. It does not write the Table file to disk, but it marks it as changed so when you exit BioEditor it will ask you to save the Table file, if you have not already done so.
Update Table Source forces BioEditor to update all its Source file pointers to the current Source directory. It does not write the Table file to disk, but it marks it as changed so when you exit BioEditor it will ask you to save the Table file, if you have not already done so.
Copyright © 2000-2010 by John Dunn and Algorithmic Arts. All Rights Reserved.